- Commonwealth Scientific and Industrial Research Organisation, Agriculture and Food, Queensland Bioscience Precinct, St Lucia, QLD, Australia
Strategies to manage metabolic hydrogen ([H]) in the rumen should be considered when reducing ruminant methane (CH4) emissions. However, little is known about the use of dietary treatments to stimulate rumen microorganisms capable of capturing the [H] available when CH4 is inhibited in vivo. The effects of the phenolic compound phloroglucinol on CH4 production, [H] flows and subsequent responses in rumen fermentation and microbial community composition when methanogenesis is inhibited were investigated in cattle. Eight rumen fistulated Brahman steers were randomly allocated in two groups receiving chloroform as an antimethanogenic compound for 21 days. Following that period one group received chloroform + phloroglucinol for another 16 days, whilst the other group received only chloroform during the same period. The chloroform treatment resulted in a decrease in CH4 production and an increase in H2 expelled with a shift in rumen fermentation toward higher levels of propionate and formate and lower levels of acetate at day 21 of treatment. Bacterial operational taxonomic units (OTUs) assigned to Prevotella were promoted whilst Archaea and Synergistetes OTUs were decreased with the chloroform treatment as expected. The shift toward formate coincided with increases in Ruminococcus flavefaciens, Butyrivibrio fibrisolvens, and Methanobrevibacter ruminantium species. The addition of chloroform + phloroglucinol in the rumen resulted in a decrease of H2 expelled (g) per kg of DMI and moles of H2 expelled per mol of CH4 decreased compared with the chloroform only treated animals. A shift toward acetate and a decrease in formate were observed for the chloroform + phloroglucinol-treated animals at day 37. These changes in the rumen fermentation profile were accompanied by a relative increase of OTUs assigned to Coprococcus spp., which could suggest this genus is a significant contributor to the metabolism of this phenolic compound in the rumen. This study demonstrates for the first time in vivo that under methanogenesis inhibition, H2 gas accumulation can be decreased by redirecting [H] toward alternative sinks through the nutritional stimulation of specific microbial groups. This results in the generation of metabolites of value for the host while also helping to maintain a low H2 partial pressure in the methane-inhibited rumen.
Introduction
Methane is a potent greenhouse gas and also represents a loss of gross feed energy for the ruminant, hence the increasing interest in strategies to manipulate CH4 production (Johnson and Johnson, 1995; Gerber et al., 2013). Archaea produce CH4 in the rumen mainly by reducing C1 compounds with H2 (among others substrates) thus maintaining a low hydrogen partial pressure (Janssen, 2010). Although methanogens are the main H2 consumers, due to the thermodynamically favorable pathway of CH4 formation, there are rumen bacteria, which are also able to use H2 as substrate thus generating alternative end products (Leng, 2014). Thus, when CH4 production is decreased, [H] might be redirected into alternative sinks nutritionally useful for the ruminant (Ungerfeld, 2015a). However, an excessive increase of the H2 partial pressure in the rumen after methane inhibition might have detrimental effects on rumen function (Wolin et al., 1997). Therefore, management of H2 accumulation in the rumen is an important strategy to consider when inhibiting CH4 emissions from livestock.
A recent published study (Martinez-Fernandez et al., 2016), using chloroform as an antimethanogenic compound, has examined the [H] flow and rumen microbial community responses in cattle fed with hay or supplemented with concentrate. It showed that a CH4 reduction of about 30% had no detrimental effects on rumen function in cattle. With both diets a decrease in CH4 production was accompanied by an increase in H2 expelled. Interestingly, the animals fed with the concentrate diet eructated relatively more H2 than the hay diet suggesting inefficient redirection of [H] into other fermentation end products. The main changes observed in rumen fermentation metabolites with both diets were a shift toward propionate and branched-chain fatty acids; an accumulation of formate, particularly with the concentrate diet; and an increase in the concentration of amino acids, organic and nucleic acids. However, although a redirection of [H] was observed, it was predicted that dietary supplements would be needed to increase the capture of excess H2 via alternative energy yielding metabolic pathways (Martinez-Fernandez et al., 2016). Failure to capture the excess H2 released from the system should be considered as an energy loss to the animal.
Microbial metabolic processes, such as reductive acetogenesis, propionogenesis, reduction of nitrate and sulfate, formate formation, and an increase of microbial biomass production when CH4 is inhibited, have been identified as nutritionally useful [H] sinks in the rumen (Newbold et al., 2005; van Zijderveld et al., 2010; Leng, 2014; Gagen et al., 2015; Ungerfeld, 2015b). Another [H] sink which has gained little attention is the reduction of phenolic compounds by rumen microorganisms. Phenolic compounds, such as flavonoids, are present in many of the forage plants consumed by ruminants. Rumen bacteria, in some cases are capable of transforming these compounds into energy yielding products for the host (Murdiati et al., 1992; McSweeney et al., 2001). Flavonoids undergoing microbial degradation in the rumen generally form phloroglucinol as an intermediate metabolite (Supplementary Figure 1). Several studies have shown that specific rumen bacteria are able to reduce phloroglucinol using H2 or formate as electron donors thus yielding acetate as the terminal product (Tsai and Jones, 1975; Patel et al., 1981; Krumholz and Bryant, 1986). However, there is scarce information about the use of unconventional dietary treatments that may stimulate rumen microorganisms capable of capturing the [H] available when CH4 is inhibited in vivo.
The aims of the present study were to analyze the effect of phloroglucinol on CH4 production, [H] flows and subsequent responses in rumen fermentation and microbial community composition in cattle when methanogenesis is inhibited. It was hypothesized that less H2 would be expelled by the chloroform + phloroglucinol-treated animals due to a shift in fermentation toward acetate by the reduction of the phenolic compound.
Materials and Methods
The experimental protocol complied with the Australian Code for the Care and Use of Animals for Scientific Purposes (eighth edition, 2013) and was approved by the local Animal Experimentation and Ethics Committee (A08/2014).
Treatments
The antimethanogen compound used was a halogenated hydrocarbon (chloroform) entrapped in a β-cyclodextrin matrix (6–7% w/w chloroform) as described by Martinez-Fernandez et al. (2016). The phenolic compound used was phloroglucinol (Thermo Fisher Scientific, Scoresby, VIC, Australia; purity of 99%).
Experimental Design and Sampling
Eight rumen fistulated Brahman (Bos indicus) steers (live weight, 365 ± 12 kg) at Lansdown Research Station (Townsville, QLD, Australia) were randomly allocated in two groups (four animals per group), receiving a diet ad libitum with a ratio 60:40 forage:concentrate (Rhodes grass hay; chemical composition: dry matter (DM), 917 g/kg fresh matter; in g/kg of DM: organic matter, 814; crude protein (CP), 109; neutral detergent fiber (NDF), 651; acid detergent fiber (ADF), 351. Concentrate; Ridley AgriProducts Pty Ltd., Brisbane, QLD, Australia. Ingredients (g/kg): barley (458), sorghum (200), molasses (30), wheat (200), legume hulls (80), urea (2.5); concentrate chemical composition: DM, 887 g/kg fresh matter; in g/kg of DM: CP, 114; NDF, 199; ADF, 130; and fat, 24). Animals were adapted to the diet over 30 days and then placed into individual pens for the measurement of individual intakes (10 days) and treated through the cannula with cyclodextrin (2 g/100 kg LW). On days 9 and 10, animals were placed into open-circuit respiration chambers for measurement of CH4 and H2 production and collection of rumen samples. Following the initial control period animals received the chloroform through the rumen-cannula, increasing the dose progressively during 21 days (treatment period 1, 1.6 g/100 kg LW, divided into two doses per day) with the last 2 days confined in open-circuit respiration chambers for direct measurement of CH4 and H2 production and rumen fluid collection. After that period, one group received the chloroform + phloroglucinol for 16 days, whilst the other group received only the chloroform treatment (treatment period 2). Phloroglucinol was placed in the rumen through the cannula twice per day, progressively increasing the dose during the first 10 days up to 75 g/100 kg LW (estimated rumen concentration 40 mM). The last 2 days of that period rumen samples and CH4 and H2 measurements were taken as previously described.
Rumen fluid samples (approximately 60 ml per animal) were collected through the cannula of the animal using a probe (covered with two layers of cheesecloth) at 3 h post feeding during confinement in respiration chambers to determine the effect on rumen fermentation parameters and rumen microbial communities. Rumen samples were stored at -20°C for short chain fatty acid (SCFA) and NH3-N analyses. Additionally, 20 ml were kept at -80°C prior to DNA extractions.
Four open circuit respiration chambers were used to determine CH4 and H2 production from individual steers as described by Martinez-Fernandez et al. (2016).
Chemical Analysis
The feed samples were dried in a forced-air oven at 65°C prior to grinding. Feed samples were ground through a 1-mm sieve before analysis. DM, ash, NDF, ADF, fat, and total nitrogen contents were analyzed by Symbio Alliance (Eight Mile Plains, QLD, Australia) following the accredited methods CF006.1, CF007, CF038.1, CF038.3, CF004, and CF003.2, respectively [AOAC International (2005) official methods: 925.10, 923.03, 920.39, 990.03, 2002.04, and 973.18]. The nitrogen values were converted to CP by a multiplication factor of 6.25.
Rumen fluid concentrations of SCFAs (acetate, propionate, n-butyrate, isobutyrate, isovalerate, and n-valerate) were measured by gas chromatography (GC) as described by Gagen et al. (2014). Isovalerate (3-methyl butyrate) includes 2-methyl butyrate, which co-elutes. The rumen NH3-N concentration was determined by the colorimetric method of Chaney and Marbach (1962).
An UltiMate®3000 HPLC system (Dionex, Sunnyvale, CA, United States) with a dedicated Photodiode Array Detector and an Autosampler was used to determine the concentration of formic acid in rumen fluid as described by Gagen et al. (2014).
Calculation of [H] Redirection and Non-carboxyl SCFA Carbons
The differences between treatments regarding incorporation of [H] into SCFA (HUSr) and formate (HUFr) were estimated using the concentrations of metabolites as a proxy since actual production rates were not measured. The stoichiometry was calculated as described by Martinez-Fernandez et al. (2016). The CH4 gas production (GP; mol/day) and H2 gas production (GP; mol/day) was used to calculate the ratio between [H] redirected to H2 expelled/CH4 decreased (Martinez-Fernandez et al., 2016).
DNA Extractions
DNA extractions from rumen samples were performed using the cetyltrimethylammonium bromide (CTAB) method of Brookman and Nicholson (2005) with minor modifications as follows: samples were centrifuged (13,000 × g for 5 min), and the supernatant was removed before DNA extraction. Cells were homogenized with 200 mg of silica-zirconium beads (1:1 mixture of 0.1- and 1.0-mm beads; Biospec, Bartlesville, OK, United States) and 800 μl of CTAB buffer in a Mini-Beadbeater-8 (Biospec) on maximum speed for 2 min, twice. Samples were incubated at 70°C for 20 min and centrifuged at 10,000 × g for 10 min, and the supernatant was mixed with 500 μl of 25:24:1 phenol:chloroform:isoamyl alcohol (Fluka BioChemika, Buchs, Switzerland).
16S rDNA and Statistical Analyses
High throughput sequencing platforms, barcoding procedures for sample recognition and phylogenetic analysis of the 16S rDNA gene were used to characterize the microbial populations present in the rumen for the control and treatment periods. The V4 region of the 16S rRNA gene was targeted using specific primers (Kozich et al., 2013). Each individual DNA sample was amplified using the specific primers and a unique barcode combination. Afterward, amplification products were visualized by performing gel electrophoresis. Product quantities were calculated and an equal molar amount of each product was pooled. The pooled products were run in a 1% agarose gel and bands were visualized and excised under blue light transillumination. The amplicons were gel purified with QIAquick Gel extraction Kit (Qiagen, Hilden, Germany) prior to submission for Illumina Miseq (Macrogen Inc., South Korea).
Paired end short read sequence data generated on the Illumina Miseq was processed using the USEARCH package (Edgar, 2010). De-multiplexed paired end sequences were first merged prior to sequence quality filtering, followed by denoising (error correction) and chimera checking and clustering of sequences to operational taxonomic units (OTUs) of 97% similarity. Taxonomic assignment of sequences was performed against the Greengenes database (McDonald et al., 2012). Additional analysis of OTUs was performed in the R packages vegan, Phyloseq and DESeq2 and the ggplot2 graphics package (McMurdie and Holmes, 2013; Oksanen et al., 2013; Love et al., 2014; Wickham, 2016). The significances of grouping in the PCoA plots were tested by analysis of dissimilarity (ADONIS) with 999 permutations. The sequences obtained in this paper have been deposited in the European Nucleotide Archive under the accession number (PRJEB20458).
The effect of treatment was analyzed for CH4/H2 production, DMI, ADWG, and rumen fermentation metabolites. Chloroform only and chloroform + phloroglucinol groups were compared as a univariate model using the GLM procedure of SPSS (IBM, version 21.0), the treatment was considered the fixed effect with the animal as experimental unit. Data from the chloroform only treatment at day 21 and their respective controls (non-treated period) were analyzed separately as a repeated-measures analysis using the linear mixed-model of SPSS (IBM, version 21.0), with the animal as the experimental unit. Effects were declared significant at P < 0.05 and P-values between 0.05 and 0.10 were considered as a trend.
Results
Ruminal Fermentation and Gas Production
No significant effects were observed on DMI, average daily weight gain, CH4 and H2 production (g/kg DMI), and fermentation parameters between experimental groups at either the control period or at day 21 of chloroform treatment before supplementation with phloroglucinol (Supplementary Tables 1, 2). A significant decrease (P ≤ 0.05) in CH4 production (g/kg DMI) (40% reduction) and increase in H2 (1.2 H2 g/kg DMI) were observed in both groups of animals treated with chloroform compared with their respective control period. The rumen fermentations parameters showed an increase (P ≤ 0.05) of propionate, branched-chain fatty acids, and formate in chloroform-treated animals compared with the control period (Supplementary Table 3).
Animals that continued to receive only chloroform continued to maintain lower CH4 emissions, but changes were observed in VFA profiles between days 21 and 37. With continued chloroform treatment there was a decrease in the molar percentage of propionate with concurrent increases in acetate and formate concentration (Table 1 and Supplementary Table 3). Conversely those animals supplemented with chloroform + phloroglucinol at day 37 had a significant (P ≤ 0.05) reduction in both the amount of H2 expelled (g) per kg of DMI and moles of H2 expelled per mol of CH4 decreased compared with the chloroform only treated animals (Table 1). Rumen fermentation parameters showed a significant increase in acetate and reduction in formate concentration (P ≤ 0.05) in animals treated with chloroform + phloroglucinol compared with the chloroform only group. Daily weight gain increased significantly (P ≤ 0.05) in chloroform + phloroglucinol treated animals compared with the chloroform only treated group (1.38 vs 0.438 kg/day, respectively) at day 37.
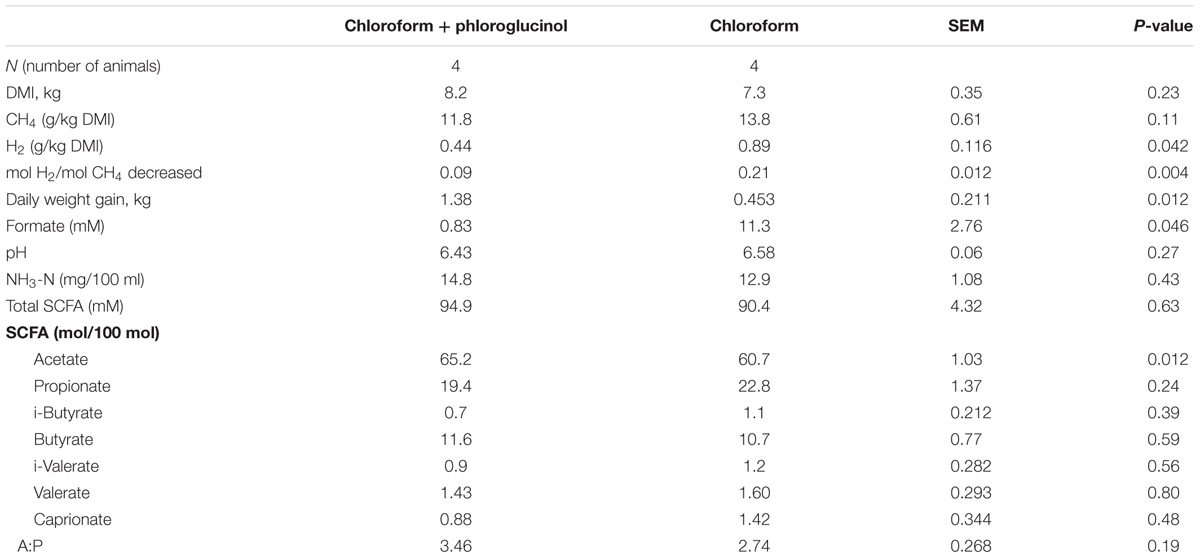
TABLE 1. Chloroform and chloroform + phloroglucinol effects on DMI, CH4, H2, daily weight gain, and rumen fermentation parameters in animals at day 37.
The calculations of [H] flows showed no significant differences in the [H] redirection into SCFA between chloroform + phloroglucinol treated animals and the chloroform only group (Figure 1A). Whereas, the chloroform + phloroglucinol treated animals showed a significant (P ≤ 0.05) decrease in [H] redirected into formate compared to the chloroform only treatment (Figure 1B). Furthermore, a significant decrease (P ≤ 0.01) of [H] redirected into H2 per mol of CH4 decreased (Figure 1C) was observed for the chloroform + phloroglucinol group compared with the chloroform only treated group.
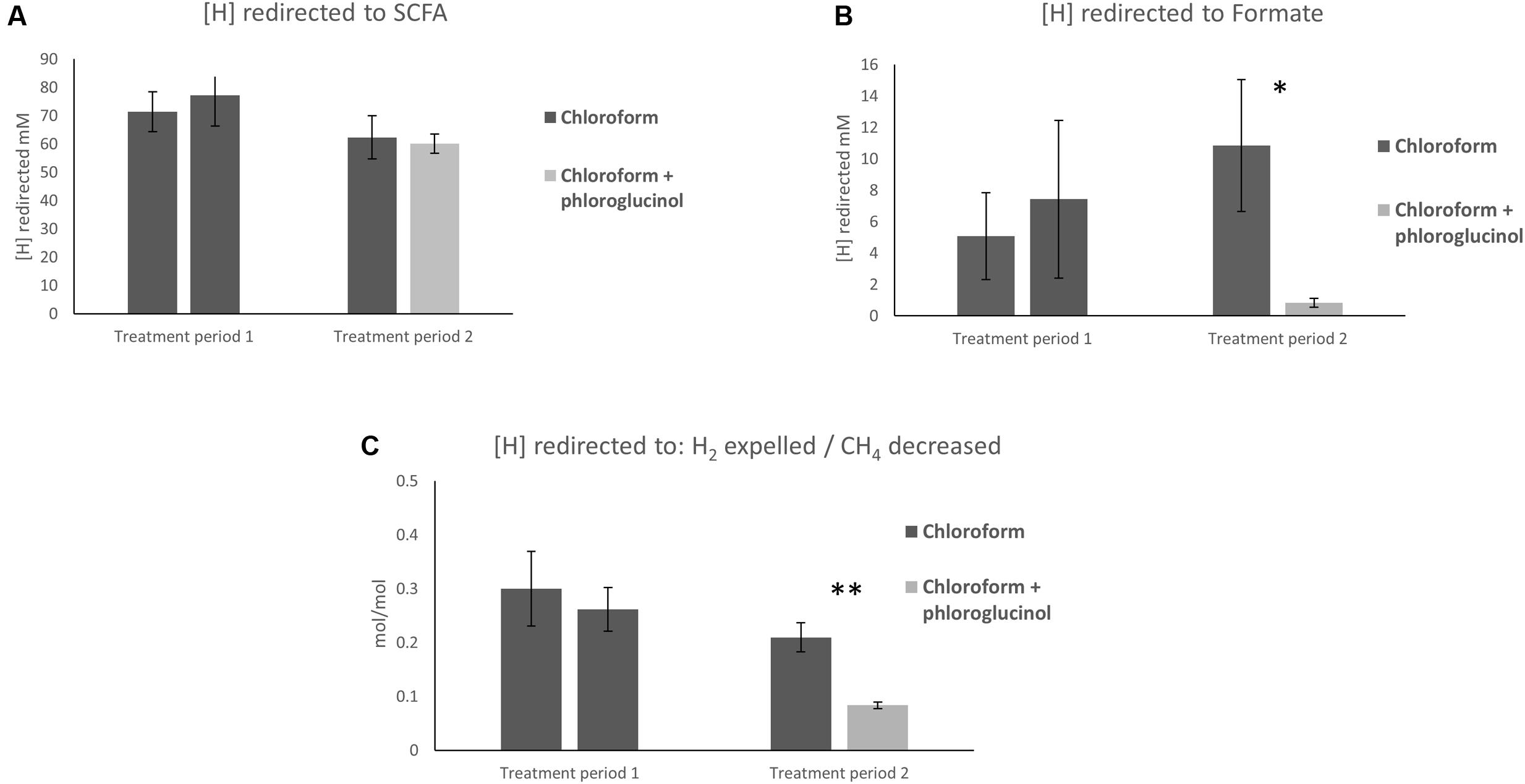
FIGURE 1. Effect of treatments on (A) redirection of [H] to SCFA, (B) formate, and (C) the ratio between [H] redirected to H2 expelled/CH4 decreased. Asterisks “∗” and “∗∗” denote significant differences between treatments, P < 0.05 and P < 0.010, respectively.
Microbial Community
The diversity analysis of the rumen microbiota showed that total microbial species richness and evenness (Shannon and Simpson, respectively) were not impacted significantly either by treatment or time (Supplementary Figure 2).
The structure of the microbiome as determined by non-phylogenetic beta diversity analysis (Bray–Curtis) showed the largest variance was due to time (treatment effect), with the control period more different than treatments at 21 and 37 days (Supplementary Figure 3). Time explained 14% of the variance from the centroid (P < 0.001), while the animal groups only explained 4% (P = 0.03), showing a separation between chloroform and chloroform + phloroglucinol treated animals at day 37 (beta dispersions; P = 0.151).
The comparison of the rumen microbiome showed a shift in specific OTUs between the chloroform treated animals and the control period, with an increase of OTUs classified in the Prevotella genus and a decrease in OTUs assigned to the Archaea domain in the chloroform animals compared with the control period (Supplementary Figure 4). Animals that continued with the chloroform only treatment to day 37 showed decreases in OTUs assigned to Prevotella and increases for those assigned with Ruminococcus, Butyrivibrio, and Methanobrevibacter (Supplementary Figures 5–7). The chloroform + phloroglucinol treatment at day 37 was associated with an increase in OTUs affiliated with the genera Prevotella, Ruminococcus, Fibrobacter, CF231, YRC22, and Coprococcus compared to animals receiving only chloroform, with OTUs assigned to Coprococcus and Prevotella showing the greatest fold change (Figure 2). Compared to chloroform only treatment, the addition of phloroglucinol altered the methanogen rank abundance back to one more representative of the control animals (Figure 3). Within the Methanobrevibacter genus, Methanobrevibacter gottschalkii and Methanomassiliicoccaceae associated OTUs remained suppressed, while Methanobrevibacter ruminantium OTUs were significantly increased on day 37 (Figure 3).
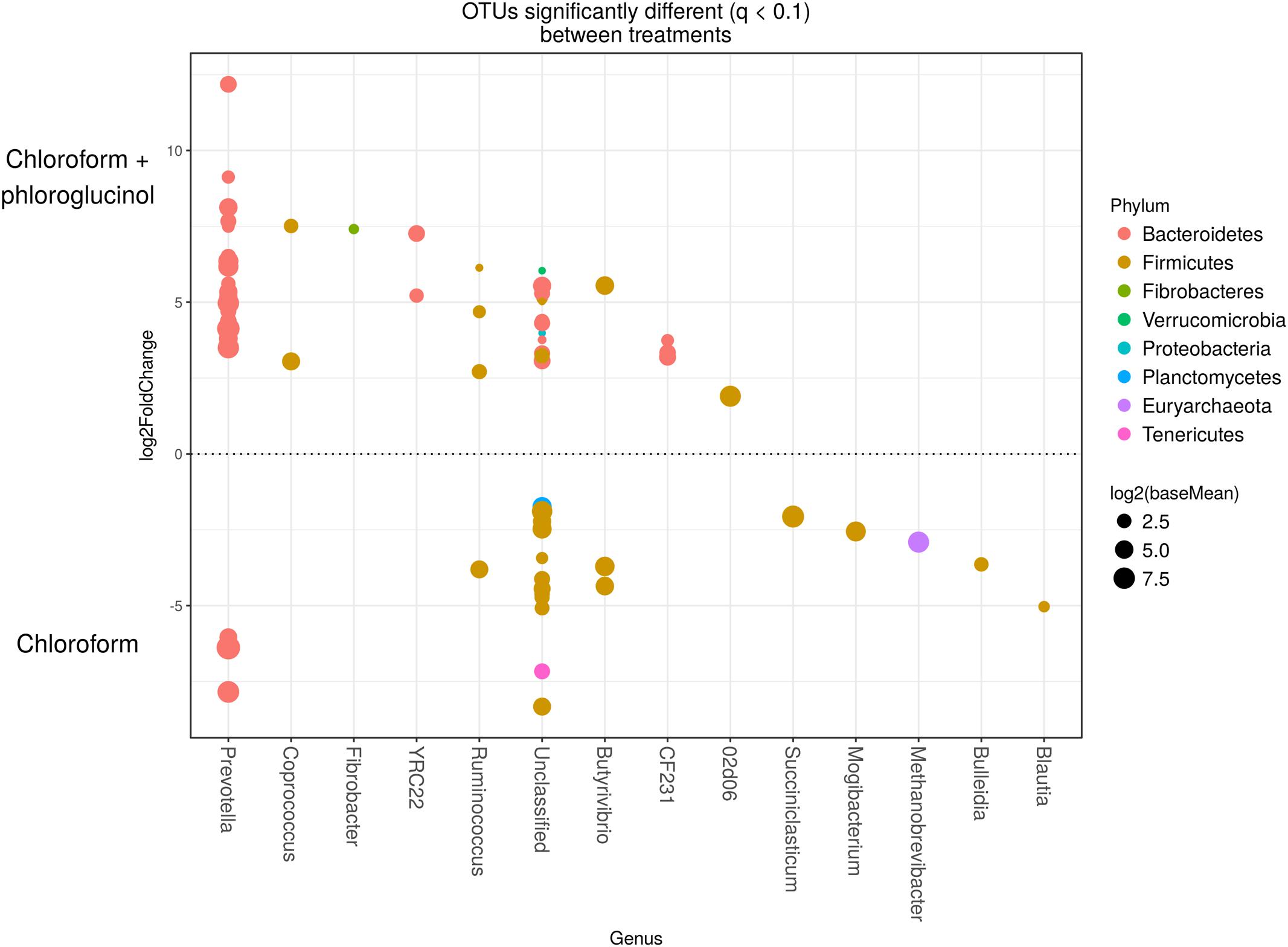
FIGURE 2. OTUs significantly different (q < 0.1 FDR) between chloroform + phloroglucinol and chloroform treated-animals. Upper axis represents OTUs with a log2 fold positive difference for chloroform + phloroglucinol treatment relative to chloroform while the lower y-axis is the negative fold difference of the chloroform + phloroglucinol relative to chloroform. Each point represents a single OTU colored by phylum and grouped on the x-axis by taxonomic genus level, size of point reflects the log2 mean abundance of the sequence data.
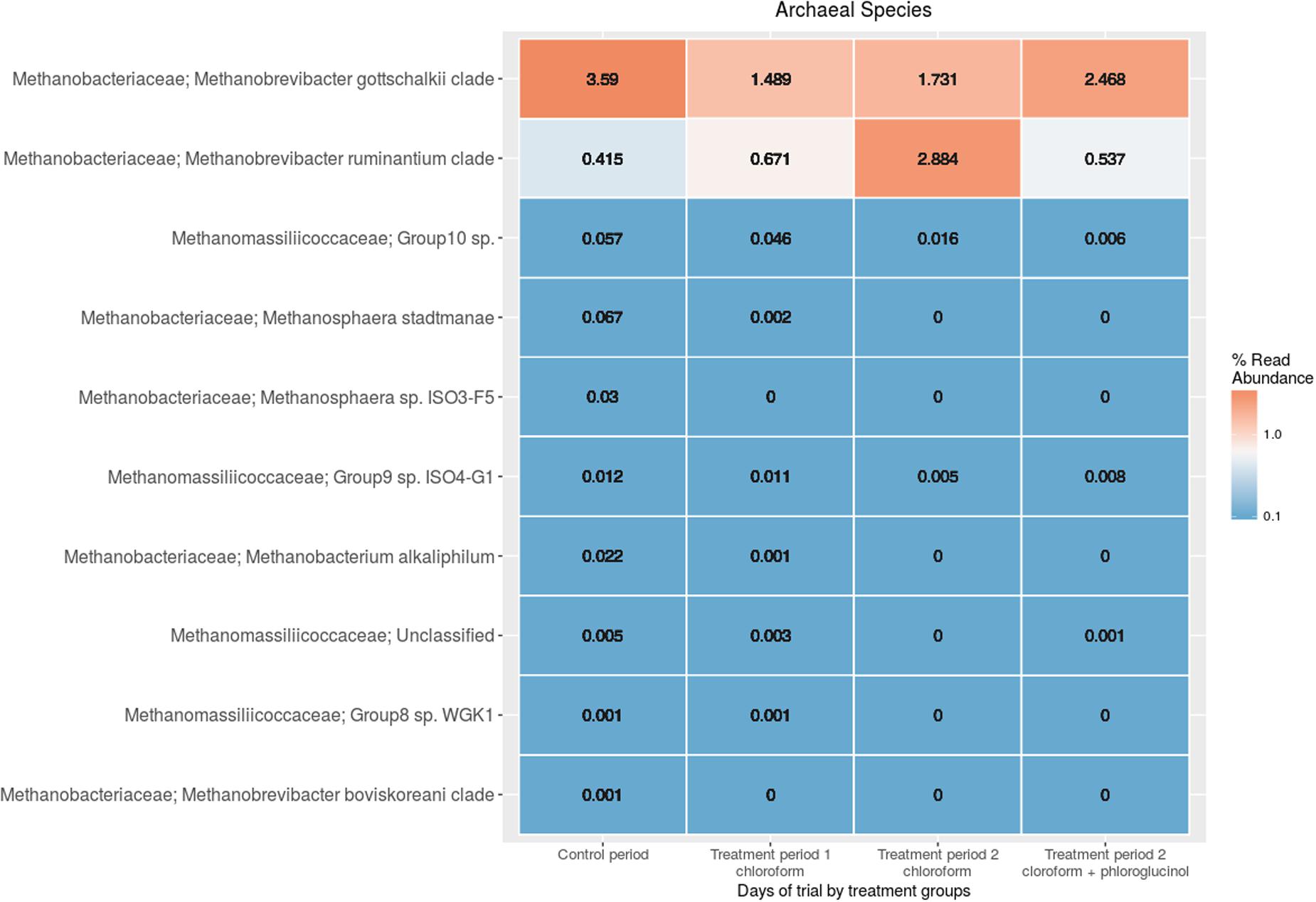
FIGURE 3. Rumen methanogen community heat maps of the most abundant OTUs using regularized log transformed values from DeSeq2 at control period, treatment period 1 and treatment period 2.
The relative abundance of the Coprococcus genus was significantly (P < 0.001) increased in the chloroform + phloroglucinol treated animals at day 37 compared with the chloroform-only treatment at day 37 and the control period (Figure 4). A significant (P < 0.001) decrease in the Synergistetes phylum was observed with chloroform and chloroform + phloroglucinol treated animals at day 37 compared with the untreated animals at day 0 (Supplementary Figure 8). Synergistetes members were also more abundant (P = 0.003) in chloroform + phloroglucinol treated animals compared with the chloroform only group at day 37.
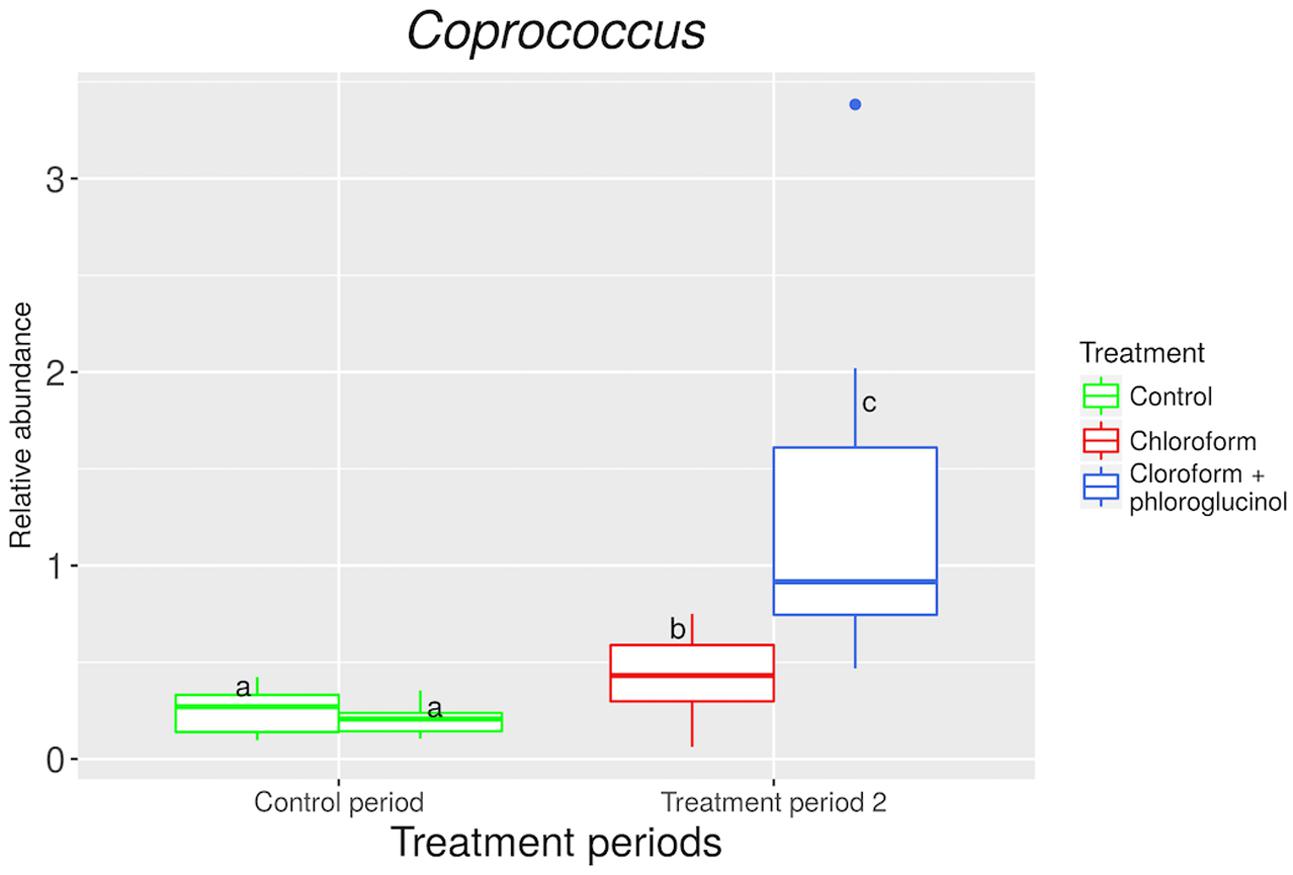
FIGURE 4. Relative abundance of OTUs assigned to Coprococcus genus at control period (untreated animals) and treatment period 2 (chloroform + phloroglucinol and chloroform-treated animals). a,bLetters denote significant differences between groups, bars that do not share the same letter are significantly different from each other (P < 0.05).
Discussion
Inhibition of methanogenesis in ruminants using halogenated compounds has previously shown a redirection of [H] toward propionate and branched-chain fatty acids with increased H2 eructation from the rumen (Mitsumori et al., 2012; Martinez-Fernandez et al., 2016). Likewise, the administration of chloroform to animals in this trial produced a similar response, thus providing a model system for testing the ability to alter rumen metabolic pathways through the capture of excess H2. A number of alternative [H] sinks have been identified for when methanogenesis is inhibited in ruminant livestock such as: acetogenesis (Joblin, 1999; Fonty et al., 2007; Gagen et al., 2015), propionogenesis (Newbold et al., 2005), nitrate reduction to ammonia (Morvan et al., 1996; Anderson et al., 2003; van Zijderveld et al., 2010; Latham et al., 2016), sulfate reduction to hydrogen sulfite (van Zijderveld et al., 2010), formate formation (Leng, 2014), and increases of microbial biomass production (Ungerfeld, 2015b). However, there is scarce information about the amount of [H] redirected to these metabolic processes or emitted as H2 gas under in vivo conditions. While the rumen microbiota can adapt to inhibition of CH4 formation by redirecting a proportion of [H] into energy-yielding metabolites, an important proportion of the unutilized [H] accumulates and is eructated as H2 gas, which still represents an energy loss to the animal. This study demonstrates for the first time in vivo that under methanogenesis inhibition, H2 gas accumulation can be reduced by redirecting [H] toward alternative sinks through the nutritional stimulation of microbial groups that generate metabolites of value to the host. Therefore, the use of dietary treatments to promote key rumen microbial groups, may be a practical way of capturing [H] that would normally be lost as H2 gas when a strategy of directly inhibiting methanogens in the rumen is employed to reduce CH4 formation.
Interestingly, an increase in daily weight gain was observed for the chloroform + phloroglucinol treated animals, indicating that the [H] redirection toward an alternative sink facilitated the utilization of a substrate, which is not normally regarded as an energy-yielding nutrient for the host. Although the increase in weight gain is a promising result, it should be interpreted with caution due to the relatively short length of the trial and small number of animals used. A recent trial with dairy cattle using 3-nitrooxypropanol to reduce CH4 emissions produced an 80% increase on the average body weight gain over the experimental period (Hristov et al., 2015). While this most likely indicates a shift in metabolism to end products of greater nutritional or metabolic value, the authors also observed a 64-fold increase in hydrogen emissions, suggesting that further efficiencies could be gained. However, to confirm this hypothesis further in vivo research on rumen digestibility and metabolism should be carried out.
In relation to the redirection of [H], the inclusion in the rumen of the phenolic substrate phloroglucinol resulted in greater acetate formation with less H2 expelled per mole of CH4 decreased, and decreased formate concentration in the rumen. This is in agreement with in vitro studies (Patel et al., 1981; Krumholz and Bryant, 1986) that have shown that rumen microorganisms were able to metabolize phloroglucinol to acetate by consuming H2 or formate as reducing agents. It should be noted that the formula for calculation of the [H] used in SCFA formation (Martinez-Fernandez et al., 2016) does not account for the acetate produced through reductive processes, such as acetogenesis or reduction of phenolic compounds, which most likely explains why there was no difference in the calculated flow of [H] into these organic acids between the treatments. Chloroform + phloroglucinol treated animals expelled per day 1.42 mol of H2 less than the chloroform only treated group, which is lower than the predicted moles of H2 (2.18) used to degrade the amount of phloroglucinol provided per day (1 molecule phloroglucinol + 1 molecule H2 = 2 molecules acetate + 2 molecules carbon dioxide (Tsai et al., 1976; Conradt et al., 2016) (Supplementary Figure 1). This discrepancy might indicate that phloroglucinol was not totally metabolized, however, near complete degradation of similar quantities of phenolic compounds have been reported previously (Murdiati et al., 1992). It is more likely that both formate and H2 were used to reduce phloroglucinol based on the lower formate concentrations in the chloroform + phloroglucinol treated animals (0.83 vs 11.3 mM) compared with chloroform only. Formate is produced during normal rumen fermentation and is mainly consumed by methanogens to produce CH4 (Hungate et al., 1970; Asanuma et al., 1998). Similar to the current study, an increase in formate has been reported previously in steers which were treated with chloroform alone to reduce methanogenesis, particularly with concentrate-supplemented diets (Martinez-Fernandez et al., 2016). Our results support the hypothesis that increased formate accumulation represents a response by the rumen microbiota to control H2 partial pressures in the rumen, by acting as a hydrogen sink (Leng, 2014; Ungerfeld, 2015b; Martinez-Fernandez et al., 2016).
Members of the Synergistetes phylum are sensitive to H2 partial pressures within the rumen and generally are reduced when the H2 partial pressure increases or methanogenesis is restricted (Denman et al., 2015; Wallace et al., 2015; Martinez-Fernandez et al., 2016). Similar reductions in the Synergistetes phylum were observed in this study with the chloroform-only treatment and increased H2 partial pressures. The addition of phloroglucinol and redirection of [H] led to lower partial pressures of H2 and probably resulted in increases in the Synergistetes phylum further supporting their significance as a key indicator group around hydrogen transactions.
Of particular interest, was the greater increase in formate concentration when the animals were exposed to longer periods of chloroform, allowing for subsequent increases in the acetate:propionate ratio, presumably driven by the decrease in H2 partial pressures. Fibrolytic Ruminococcus species are able to alter their fermentative pathways to provide alternative reducing equivalent sinks, including the production of formate and ethanol to relieve the inhibition of the reoxidation of NADH (Miller and Wolin, 1974; Shi et al., 1997). With the higher partial pressures of H2 observed in the rumen for the chloroform treated animals, the shift to increased formate might be linked with increases of Ruminococcus flavefaciens, Butyrivibrio fibrisolvens, and M. ruminantium species. Methanogen species capable of utilizing formate are also able to produce formate from CO2 and H2 under conditions of high H2 partial pressures and when inhibited with halogenated hydrocarbons (Thiele and Zeikus, 1988; Bleicher and Winter, 1994). The observed increase in the relative abundance of M. ruminantium over the predominant rumen methanogen M. gottschalkii, which is incapable of formate utilization, likely reflects the activation of this mechanism. By converting H2 and CO2 to formate while H2 is in excess and methanogenesis is impeded may ensure survival until conditions become more favorable for producing CH4, thus providing M. ruminantium with a competitive advantage over non-formate utilizing methanogens. The addition of phloroglucinol altered these conditions by redirecting [H] away from formate and reducing the H2 partial pressure which allowed M. gottschalkii to outcompete M. ruminantium and once again become the dominant methanogen species in this system. The changes in the microbiota illustrate the ability of the rumen to approach a new equilibrium by adapting to the altered environmental conditions. Of course, this also means that these changes might not correspond to a fully adapted and stable rumen microbiome as is illustrated by the continued shift in the microbiome for the chloroform only group between days 21 and 37. Longer trial periods would need to be studied to fully answer if the rumen has reached a stable microbiota.
Phloroglucinol is a phenolic compound, formed through the degradation of complex plant molecules such as flavonoids, which are commonly present in the diet of the grazing ruminant (Tsai et al., 1976). It is also present at high levels in brown algae (Ecklonia cava) which is widely harvested for its medicinal effects (Park et al., 2012). In the rumen, microorganisms degrade this compound predominately to produce acetate and CO2 as end-products. Our findings are in accordance with previous in vitro studies (Tsai et al., 1976; Patel et al., 1981; Krumholz and Bryant, 1986), which observed this increase in acetic acid concentration when phloroglucinol was metabolized by rumen microorganisms. Several rumen bacteria have been identified as phloroglucinol degraders and were classified as Eubacterium oxidoreducens, Streptococcus bovis, and Coprococcus spp. (Tsai et al., 1976; Patel et al., 1981; Krumholz and Bryant, 1986). Krumholz and Bryant (1986) reported that E. oxidoreducens required H2 or formate to degrade phloroglucinol to acetate. The analysis of the rumen microbiome in this study did not identify E. oxidoreducens but did reveal several OTUs assigned to Coprococcus spp. increased within the chloroform + phloroglucinol treated animals, suggesting the genus is a significant contributor to the metabolism of this phenolic compound. Tsai et al. (1976) found that Coprococcus spp. were able to degrade 1 molecule of phloroglucinol to 2 molecules of acetic acid and 2 molecules of carbon dioxide. A more detailed study by Patel et al. (1981) showed that phloroglucinol is initially reduced to dihydrophloroglucinol by Coprococcus spp. using NADPH as the electron donor. This would help alleviate the increase in NADPH in the rumen generated by the inhibition of methanogenesis through its oxidation to NADP+ (Chalupa, 1977). Furthermore, a recently published study (Conradt et al., 2016) identified three phloroglucinol reductases belonging to the family of NADPH dehydrogenases/reductases involved in the anaerobic degradation of phloroglucinol which involves hydrolytic ring cleavage to 3-hydroxy-5-oxohexanoic and then acetate formation. However, to confirm our hypothesis, further studies using metagenomics and transcriptomic approaches should be carried out to evaluate the functional genes in Coprococcus spp. associated with the NADPH-dependent reduction of phloroglucinol and the redirection of H2 in the rumen. Furthermore, as the limitations of the trial did not allow for the inclusion of a phloroglucinol only treatment, we cannot ascertain if these effects may have also been possible in the presence of functional methanogenesis and lower H2 partial pressures. Indeed, a calculated ΔG° of -158 kJ/mol phloroglucinol to acetate indicates this reaction is likely to be very favorable thermodynamically and may even occur in the presence of hydrogenotrophic methane formation (Kaiser and Hanselmann, 1982; Krumholz and Bryant, 1986).
The Coprococcus genus is also involved in other important metabolic pathways in the rumen. Recently, Shabat et al. (2016) found a greater proportion of Coprococcus species (Coprococcus catus) in the rumen of dairy cows with lower CH4 emissions and higher feed efficiencies. In the more feed efficient cows there were an increase in abundance of genes aligned to the acrylate pathway, which involves the conversion of lactate to propionate, and were annotated as C. catus. In general, the acrylate pathway rather than the succinate pathway for propionate production was more dominant in the efficient animals (Shabat et al., 2016). Other metabolic pathways in which Coprococcus spp. are also involved, is the degradation of nitrotoxins in the rumen (Majak and Cheng, 1981) through reductive processes. These and our findings suggest that Coprococcus genus is involved in key metabolic pathways in the rumen, and they should be considered when developing strategies to promote microorganisms which might improve the rumen efficiency and metabolize plant toxins in ruminants.
Conclusion
The present study in cattle demonstrated that under anti-methanogenesis conditions the addition of phloroglucinol into the rumen can stimulate microbial groups which utilize H2 and formate as reductants in the metabolism of phloroglucinol to acetate and thus decrease the partial pressure of H2 in the rumen and the amount of this gas eructated by the animal. This demonstrates that the negative impact of H2 accumulation in the rumen under suppression of methanogenesis can be ameliorated by the provision of novel compounds that have nutritional value for the animal when degraded by reductive processes.
Author Contributions
CM, SD, and GM-F conceived and designed the experiments and analytical approaches. GM-F performed the animal trial. GM-F and JC analyzed the biological samples. SD and GM-F analyzed the data. GM-F, CM, and SD wrote the manuscript. All authors agree to be accountable for all aspects of the work.
Funding
This research was funded by a grant from Meat and Livestock Australia and Department of Agriculture, Fisheries and Forestry, Australian Government (MLA- B CCH 7610; Department of Agriculture 1194246–108).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We gratefully acknowledge S. Austin for his assistance with the animals.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01871/full#supplementary-material
References
Anderson, R. C., Callaway, T. R., Van Kessel, J. A. S., Jung, Y. S., Edrington, T. S., and Nisbet, D. J. (2003). Effect of select nitrocompounds on ruminal fermentation: an initial look at their potential to reduce economic and environmental costs associated with ruminal methanogenesis. Bioresour. Technol. 90, 59–63. doi: 10.1016/S0960-8524(03)00086-5
Asanuma, N., Iwamoto, M., and Hino, T. (1998). Formate metabolism by ruminal microorganisms in relation to methanogenesis. Anim. Sci. Technol. 69, 576–584. doi: 10.2508/chikusan.69.576
Bleicher, K., and Winter, J. (1994). Formate production and utilization by methanogens and by sewage sludge consortia-interference with the concept of interspecies formate transfer. Appl. Microbiol. Biotechnol. 40, 910–915. doi: 10.1007/BF00173998
Brookman, J. L., and Nicholson, M. J. (2005). “Anaerobic fungal populations,” in Methods in Gut Microbial Ecology for Ruminants, eds H. P. S. Makkar and C. S. McSweeney (Dordrecht: Springer), 139–150.
Chalupa, W. (1977). Manipulating rumen fermentation. J. Anim. Sci. 45, 585–599. doi: 10.2527/jas1977.453585x
Chaney, A. L., and Marbach, E. P. (1962). Modified reagents for determination of urea and ammonia. Clin. Chem. 8, 130–132.
Conradt, D., Hermann, B., Gerhardt, S., Einsle, O., and Müller, M. (2016). Biocatalytic properties and structural analysis of phloroglucinol reductases. Angew. Chem. Int. Ed. Engl. 128, 15760–15763. doi: 10.1002/anie.201607494
Denman, S. E., Martinez Fernandez, G., Shinkai, T., Mitsumori, M., and Mcsweeney, C. S. (2015). Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analogue. Front. Microbiol. 6:1087. doi: 10.3389/fmicb.2015.01087
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fonty, G., Joblin, K., Chavarot, M., Roux, R., Naylor, G., and Michallon, F. (2007). Establishment and development of ruminal hydrogenotrophs in methanogen-free lambs. Appl. Environ. Microbiol. 73, 6391–6403. doi: 10.1128/AEM.00181-07
Gagen, E. J., Denman, S. E., and Mcsweeney, C. S. (2015). “Acetogenesis as an alternative to methanogenesis in the rumen,” in Livestock Production and Climate Change, eds P. K. Malik, R. Bhatta, J. Takahshi, R. A. Kohn, and C. S. Prasad (Wallingford: CABI), 292–303.
Gagen, E. J., Wang, J. K., Padmanabha, J., Liu, J., De Carvalho, I. P. C., Liu, J. X., et al. (2014). Investigation of a new acetogen isolated from an enrichment of the tammar wallaby forestomach. BMC Microbiol. 14:314. doi: 10.1186/s12866-014-0314-3
Gerber, P., Hristov, A., Henderson, B., Makkar, H., Oh, J., Lee, C., et al. (2013). Technical options for the mitigation of direct methane and nitrous oxide emissions from livestock: a review. Animal 7, 220–234. doi: 10.1017/S1751731113000876
Hristov, A. N., Oh, J., Giallongo, F., Frederick, T. W., Harper, M. T., Weeks, H. L., et al. (2015). An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc. Natl. Acad. Sci. U.S.A. 112, 10663–10668. doi: 10.1073/pnas.1504124112
Hungate, R., Smith, W., Bauchop, T., Yu, I., and Rabinowitz, J. (1970). Formate as an intermediate in the bovine rumen fermentation. J. Bacteriol. 102, 389–397.
Janssen, P. H. (2010). Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 160, 1–22. doi: 10.1016/j.anifeedsci.2010.07.002
Joblin, K. N. (1999). Ruminal acetogens and their potential to lower ruminant methane emissions. Aust. J. Agric. Res. 50, 1307–1314. doi: 10.1071/AR99004
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Kaiser, J. P., and Hanselmann, K. W. (1982). Fermentative metabolism of substituted monoaromatic compounds by a bacterial community from anaerobic sediments. Arch. Microbiol. 133, 185–194. doi: 10.1007/BF00414999
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Krumholz, L. R., and Bryant, M. P. (1986). Eubacterium-Oxidoreducens Sp-Nov requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin. Arch. Microbiol. 144, 8–14. doi: 10.1007/BF00454948
Latham, E. A., Anderson, R. C., Pinchak, W. E., and Nisbet, D. J. (2016). Insights on alterations to the rumen ecosystem by nitrate and nitrocompounds. Front. Microbiol. 7:228. doi: 10.3389/fmicb.2016.00228
Leng, R. A. (2014). Interactions between microbial consortia in biofilms: a paradigm shift in rumen microbial ecology and enteric methane mitigation. Anim. Prod. Sci. 54, 519–543. doi: 10.1071/AN13381
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Majak, W., and Cheng, K.-J. (1981). Identification of rumen bacteria that anaerobically degrade aliphatic nitrotoxins. Can. J. Microbiol. 27, 646–650. doi: 10.1139/m81-099
Martinez-Fernandez, G., Denman, S. E., Yang, C. L., Cheung, J. E., Mitsumori, M., and Mcsweeney, C. S. (2016). Methane inhibition alters the microbial community, hydrogen flow, and fermentation response in the rumen of cattle. Front. Microbiol. 7:1122. doi: 10.3389/fmicb.2016.01122
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., Desantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
McSweeney, C., Palmer, B., Mcneill, D., and Krause, D. (2001). Microbial interactions with tannins: nutritional consequences for ruminants. Anim. Feed Sci. Technol. 91, 83–93. doi: 10.1016/S0377-8401(01)00232-2
Miller, T. L., and Wolin, M. (1974). A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27, 985–987.
Mitsumori, M., Shinkai, T., Takenaka, A., Enishi, O., Higuchi, K., Kobayashi, Y., et al. (2012). Responses in digestion, rumen fermentation and microbial populations to inhibition of methane formation by a halogenated methane analogue. Br. J. Nutr. 108, 482–491. doi: 10.1017/S0007114511005794
Morvan, B., Rieu-Lesme, F., Fonty, G., and Gouet, P. (1996). In vitro interactions between rumen H2-producing cellulolytic microorganisms and H2-utilizing acetogenic and sulfate-reducing bacteria. Anaerobe 2, 175–180. doi: 10.1006/anae.1996.0023
Murdiati, T. B., Mcsweeney, C. S., and Lowry, J. B. (1992). Metabolism in sheep of gallic acid, tannic-acid and hydrolyzable tannin from Terminalia oblongata. Aust. J. Agric. Res. 43, 1307–1319. doi: 10.1071/AR9921307
Newbold, C. J., Lopez, S., Nelson, N., Ouda, J. O., Wallace, R. J., and Moss, A. R. (2005). Propionate precursors and other metabolic intermediates as possible alternative electron acceptors to methanogenesis in ruminal fermentation in vitro. Br. J. Nutr. 94, 27–35. doi: 10.1079/BJN20051445
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R., et al. (2013). Package ‘vegan’. Community Ecology Package, Version 2.
Park, E. Y., Kim, E. H., Kim, M. H., Seo, Y. W., Lee, J. I., and Jun, H. S. (2012). Polyphenol-rich fraction of brown alga Ecklonia cava collected from Gijang, Korea, reduces obesity and glucose levels in high-fat diet-induced obese mice. Evid. Based Complement. Alternat. Med. 2012:418912. doi: 10.1155/2012/418912
Patel, T. R., Jure, K. G., and Jones, G. A. (1981). Catabolism of phloroglucinol by the rumen anaerobe Coprococcus. Appl. Environ. Microbiol. 42, 1010–1017.
Shabat, S. K. B., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Miller, M. E. B., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Shi, Y., Odt, C. L., and Weimer, P. J. (1997). Competition for cellulose among three predominant ruminal cellulolytic bacteria under substrate-excess and substrate-limited conditions. Appl. Environ. Microbiol. 63, 734–742.
Thiele, J. H., and Zeikus, J. G. (1988). Control of interspecies electron flow during anaerobic digestion: significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl. Environ. Microbiol. 54, 20–29.
Tsai, C. G., Gates, D. M., Ingledew, W. M., and Jones, G. A. (1976). Products of anaerobic phloroglucinol degradation by Coprococcus Sp Pe15. Can. J. Microbiol. 22, 159–164. doi: 10.1139/m76-022
Tsai, C. G., and Jones, G. A. (1975). Isolation and identification of rumen bacteria capable of anaerobic phloroglucinol degradation. Can. J. Microbiol. 21, 794–801. doi: 10.1139/m75-117
Ungerfeld, E. M. (2015a). Limits to dihydrogen incorporation into electron sinks alternative to methanogenesis in ruminal fermentation. Front. Microbiol. 6:1272. doi: 10.3389/fmicb.2015.01272
Ungerfeld, E. M. (2015b). Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: a meta-analysis. Front. Microbiol. 6:37. doi: 10.3389/fmicb.2015.00037
van Zijderveld, S. M., Gerrits, W. J. J., Apajalahti, J. A., Newbold, J. R., Dijkstra, J., Leng, R. A., et al. (2010). Nitrate and sulfate: effective alternative hydrogen sinks for mitigation of ruminal methane production in sheep. J. Dairy Sci. 93, 5856–5866. doi: 10.3168/jds.2010-3281
Wallace, R. J., Rooke, J. A., Mckain, N., Duthie, C.-A., Hyslop, J. J., Ross, D. W., et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:839. doi: 10.1186/s12864-015-2032-0
Keywords: rumen, microbial community, phloroglucinol, CH4, H2, 16S sequencing
Citation: Martinez-Fernandez G, Denman SE, Cheung J and McSweeney CS (2017) Phloroglucinol Degradation in the Rumen Promotes the Capture of Excess Hydrogen Generated from Methanogenesis Inhibition. Front. Microbiol. 8:1871. doi: 10.3389/fmicb.2017.01871
Received: 27 June 2017; Accepted: 13 September 2017;
Published: 05 October 2017.
Edited by:
Emilio M. Ungerfeld, Instituto de Investigaciones Agropecuarias INIA – Carillanca, ChileReviewed by:
Wang Min, Institute of Subtropical Agriculture, ChinaDiego P. Morgavi, INRA Centre Auvergne – Rhône-Alpes, France
Copyright © 2017 Martinez-Fernandez, Denman, Cheung and McSweeney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gonzalo Martinez-Fernandez, gonzalo.martinezfernandez@csiro.au
 Gonzalo Martinez-Fernandez
Gonzalo Martinez-Fernandez Stuart E. Denman
Stuart E. Denman Jane Cheung
Jane Cheung Christopher S. McSweeney
Christopher S. McSweeney